Sushant Patil

Sushant Patil
Characterizing next-generation sequencing (NGS) platforms for multiplexed biosensing and aptamer discovery: G-quadruplex aptamers as a case study
Abstract:
Aptamers are single-stranded DNA or RNA molecules forming unique three dimensional structures that can bind to their targets (generally proteins or metabolites or other nucleic acids) with high specificity and affinity. Screening for aptamers requires filtering 1012 possibilities and enriching for successful binding partners– using high throughput sequencing platforms to sort out the possibilities in a single step presents clear advantages. However, aptamers are highly structured and DNA polymerases are known to have trouble processing structures in vitro. A prominent feature of several of the best-characterized aptamers is a planar structure called G-quadruplex (GQ); these structures can stack and are particularly stable. Far from being a purely synthetic construct, GQs are prevalent in many genomes, as well, and their involvement in modulating various cellular processes has been demonstrated. Before using NGS platforms for aptamer characterization it is important to find out whether NGS platforms sequence through and accurately report the location and neighboring sequence of GQs. We compared the performance of the two most popular NGS platforms, Ion Torrent and Illumina MiSeq, in sequencing systematically varied GQ templates: each platform has difficulty with templates whose melting temperature is above the instrument operating temperature. Each uses a different DNA polymerase and chemistries, so other factors may contribute. Only with the Ion Torrent is it possible to readily manipulate the reaction conditions – the addition of single stranded binding protein improved but did not eliminate the accumulated base calling errors and low sequence quality of GQ containing templates. The Ion Torrent Hi-Q Sequencing Kit did not significantly alter the results: the level of base calling error remained essentially the same or increased in some cases. Further experiments suggest that the Ion Torrent signal acquisition and processing pipeline probably fails to capture and integrate the signal from a slower than expected polymerase rate. The alternate hypothesis, of replication slippage, was tested and discounted. A potential molecular biology approach to resolving the problem has been proposed that creates a double-stranded template that the polymerase can consistently process through and the resulting slower, yet consistent, nucleotide incorporation rate can be accounted for in signal processing pipeline.
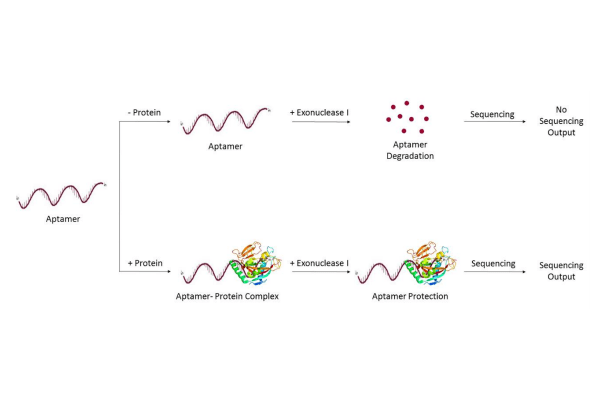
Given a suitable NGS platform that is resilient to DNA structure, the end goal is to develop a protocol for multiplexed biosensing: here we propose an experiment utilizing the principle of an exonuclease I protection assay, with case studies based on characterized a thrombin protein-binding aptamer and an acetylated histone H4 peptide-binding aptamer.
Sponsoring Chair: Dr. Jennifer Weller
Committee: Dr. Jerry Troutman, Dr. Joanna Krueger, Dr. Ed Stokes, Dr. Valery Grdzelishvili